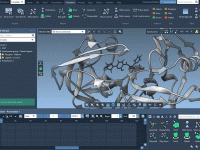
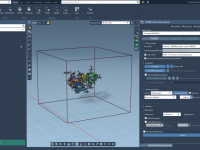
Keeping Your Focus: Track Molecular Changes Without Moving the Camera
Speed Up Your Molecular Visualization Workflow with Custom Visual Presets
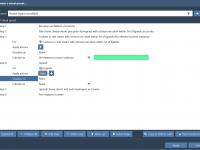
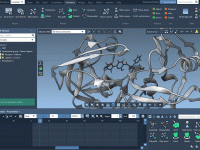
For many molecular modelers, producing high-quality visualizations from complex molecular systems is a time-consuming task. Whether for presentations, publications, or simply to better understand molecular structures, styling a model typically involves many steps — selecting components, choosing representations, applying color…
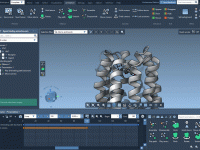
Working with Multiple Protein Replicas in Coarse-Grained Modeling
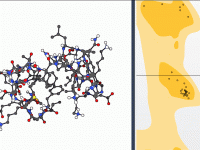
Moving Residues to Safer Ground: Interactive Refinement Using the Ramachandran Plot
Easily Control What’s Visible in Molecular Animations with the ‘Shown’ Effect
Effortless Molecular Filtering: Using NSL in the Document View
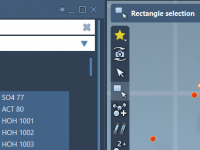
Filtering and analyzing molecular structures can become time-consuming when dealing with large assemblies. For computational chemists, structural biologists, or anyone routinely working with complex molecular data, pinpointing specific residues, bonds, chains, or functional groups quickly is essential—but often tedious. Fortunately,…
Running GROMACS Simulations in the Cloud with SAMSON
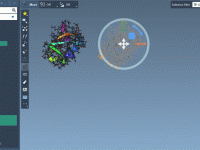
Toggling Molecular Model Visibility with NSL Made Simple
Making Smooth Molecular Flythroughs with Keyframes in SAMSON
Creating engaging molecular animations can be time-consuming, especially when trying to guide the viewer’s attention across complex molecular structures. One of the common frustrations among structural biologists, medicinal chemists, and molecular modelers is achieving smooth camera movements that emphasize the…