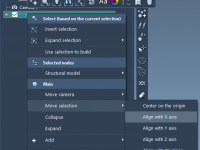
Quickly Align Molecular Structures in SAMSON with Context Menus and Compass Tools
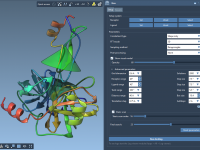
Making Sense of Side Chain Visibility in Your Molecular Models
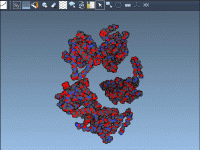
A Quicker Way to Visualize Biological Assemblies from PDB Files
Easily Show or Hide Labels in SAMSON with NSL
Improving Protein Docking Speed by Restricting the Search Domain in Hex
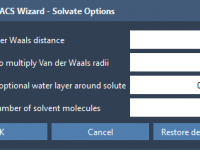
Avoiding Solvent Clashes in Coarse-Grained Simulations with MARTINI
Avoid Common Errors When Selecting Input Files in NVT Equilibration
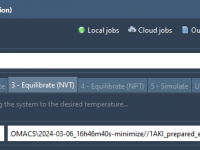
One of the subtle yet persistent challenges molecular modelers face during molecular dynamics simulations is input file mismanagement—especially during equilibration steps. When performing NVT Equilibration (constant Number of particles, Volume, Temperature), even a small mistake in choosing the right input…