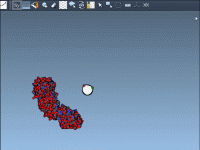
Clarifying Protein Assemblies: Preview and Generate Symmetry Mates in a Click
A Clean Way to Filter Notes in SAMSON Using NSL
From Conformations to Transition Paths: A Practical Guide to the NEB Method in SAMSON
From Zero to Simulating: Installing SAMSON Without Admin Rights
Quickly Identify Charged Regions in Molecular Models Using Structural Group Attributes
What Makes SAMSON Editors So Useful for Molecular Modeling Tasks?
From SMILES to 3D Structures in Seconds with SAMSON
Drawing Carbon Nanotubes by Hand: Quick Interactive Modeling in SAMSON

Building carbon nanotube (CNT) models is often a central need in molecular modeling, nanoscience, and materials simulation. Whether you’re designing CNT-based sensors, creating nanostructures for simulations, or exploring their mechanical and electronic behavior, the ability to quickly and intuitively generate…
Speed Up Protein Structure Predictions with AlphaFold-2 in SAMSON
Predicting accurate protein structures remains one of the most time-consuming steps in molecular modeling and drug discovery. Researchers often struggle with limited local hardware, complex pipelines, and long waiting times using public services. Fortunately, SAMSON offers an integrated and user-friendly…