How to Quickly Find Conformations with Over 100 Atoms Using NSL
A Better Way to Frame Molecules in Your Animations
Working Efficiently in SAMSON: Copy Python Code from Commands and Presets
From Raw Structure to Insight: Visual Presets in SAMSON
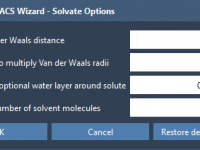
Why Solvating Coarse-Grained Systems in GROMACS Wizard Requires Extra Attention
Visualize Ligand Unbinding More Clearly with Pathlines in SAMSON
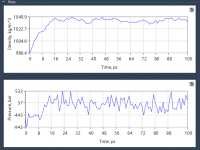
Why Your Molecular System Isn’t Ready Yet: The Importance of NPT Equilibration
How to Make Your Molecular Camera Follow the Action
Reducing Setup Time with Custom .mdp File Import in SAMSON’s GROMACS Wizard
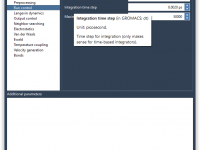
Molecular modelers working with GROMACS know the importance—and the complexity—of setting the right molecular dynamics parameters. Whether it’s energy minimization, NVT/NPT equilibration, or production simulations, configuring .mdp (molecular dynamics parameter) files can become a time-consuming part of the workflow, especially…