Reducing Setup Time with Custom .mdp File Import in SAMSON’s GROMACS Wizard
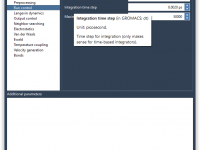
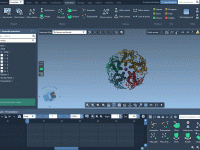
Molecular modelers working with GROMACS know the importance—and the complexity—of setting the right molecular dynamics parameters. Whether it’s energy minimization, NVT/NPT equilibration, or production simulations, configuring .mdp (molecular dynamics parameter) files can become a time-consuming part of the workflow, especially…