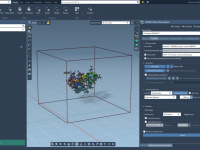
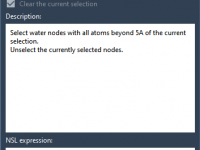
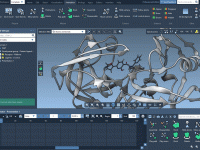
Preserving Functional Waters While Cleaning Up Your Structure
Pause, Explain, Proceed: Controlling Presentation Flow with Stop Animation in SAMSON
Whether you’re demonstrating a molecular mechanism to students or showcasing your latest simulation to collaborators, clarity matters. One underrated, yet important challenge in molecular presentations is controlling the pace at which your audience consumes dense visual information. Especially during complex…
Will SAMSON Run on Your Machine? Here’s What You Need to Know
Simplifying Molecular Arrangement: How to Align and Distribute Structures in SAMSON
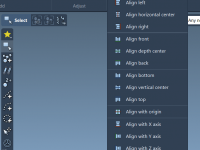
Organizing molecular structures spatially is a routine yet critical part of molecular modeling. Whether you’re preparing systems for simulations, illustrating interactions between molecules, or assembling complex biomolecular environments, correctly aligning and distributing molecules can prevent downstream errors and enhance clarity…
Lost in Molecules? Here’s How to Easily Navigate 3D Structures in SAMSON
When starting with molecular modeling, one challenge that many researchers face is navigating complex 3D structures efficiently. Whether you’re analyzing a protein-ligand interaction or building nanostructures, getting lost in space—or worse, losing your molecule entirely—happens more often than you might…