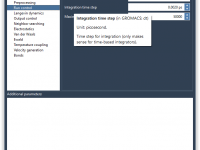
Why GROMACS Parameters Sometimes Don’t Show Up — And What To Do About It
Taming Complexity: Use Visibility Attributes to Focus Your Molecular Animations
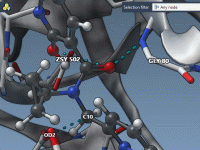
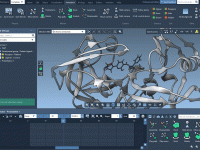
When working with complex molecular models involving multiple animations, clarity often becomes the first casualty. Every molecular modeler has experienced the frustration of an overcrowded scene—dozens of animations running simultaneously, obscuring the very phenomenon you’re trying to observe or demonstrate.…