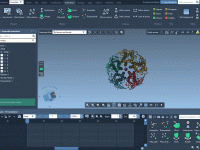
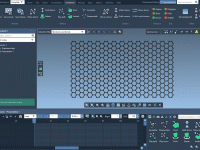
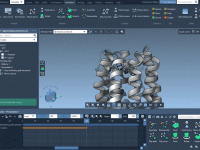
Avoiding Solvent Clashes in Coarse-Grained Simulations with GROMACS Wizard
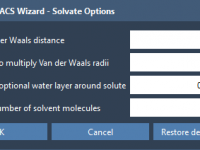
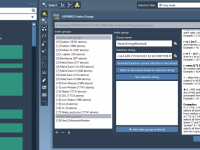
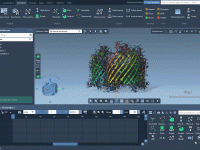
When setting up coarse-grained (CG) systems for molecular dynamics simulations with GROMACS, one common source of instability and frustration is poor solvation—especially when solvent molecules are placed too close to solute atoms, causing steric clashes. Fortunately, SAMSON’s GROMACS Wizard offers…