Making Molecules Appear—and Disappear—on Cue
Exporting DNA nanostructures to oxDNA: when design meets simulation
What Exactly Do Visual Models Do in SAMSON?
Quickly Find Molecules with the Right Structural Properties Using NSL
Keep Your Focus: Following Molecular Regions Without Moving the Camera
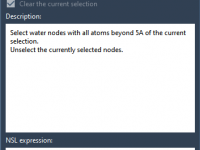
Preserving Functional Waters During Preprocessing in GROMACS Wizard
Bringing Molecular Models to Life with the Pulse Animation
Making Molecular Presentations Clearer with Orbit Camera Animations
One of the frequent challenges in molecular modeling is effectively communicating 3D structures and interactions to colleagues, students, or collaborators. Screenshots and static views are often not enough, especially when trying to showcase binding pockets, active sites, or conformational changes.…