Avoid Sampling Pitfalls: How to Set Up a Ligand for Pathway Discovery
Filtering Structural Groups by Partial Charge in SAMSON
When working with molecular models and simulations, it’s often important to quickly locate molecular fragments with specific electronic characteristics. One common use case: identifying structural groups with a particular range of partial charges — whether for reactivity analysis, parameter tuning,…
Previewing Protein Symmetry in Real Time Without Committing
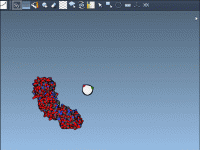
When working with crystallographic or biological assemblies of proteins, understanding how subunits are arranged is essential. Whether you’re modeling quaternary structures or designing symmetric nanostructures, replicating and visualizing symmetric interfaces can be a key step. But generating all symmetry mates…
Setting Up a Coarse-Grained MD System in Seconds: A Practical Guide for Soluble Proteins
A Simple Trick to Highlight Molecular Structures in Motion
Communicating a molecular mechanism often means going beyond static images. Whether you’re presenting a conformational change, a binding process, or a protein-ligand interaction, being able to illustrate spatial structure clearly is essential. One common challenge? Providing viewers (or yourself) with…
Progressively Hiding Atoms to Clarify Molecular Presentations
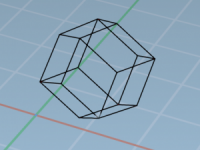
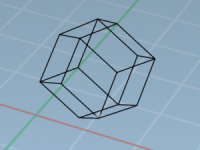
Which Unit Cell Shape Should You Use? A Toolkit for Smarter Simulations
Making Molecular Objects Appear and Disappear with Flash Animation in SAMSON
Installing SAMSON Without Admin Rights: A Relief for Molecular Modelers
Many molecular researchers experience a common yet frustrating obstacle: installing advanced molecular modeling tools on systems where administrative permissions are limited or completely restricted—especially in academic institutions, shared labs, or virtual environments. If you’ve ever faced such a hurdle, you’ll…