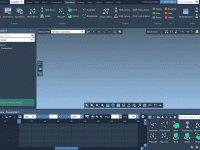
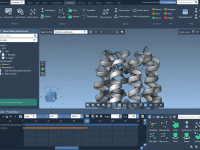
Making Molecular Structures Pop: Using Ambient Occlusion in SAMSON
Quickly Creating Custom Index Groups in GROMACS with SAMSON
Saving Molecular Animations as Movies in SAMSON
How to Identify Chemical Patterns in SMILES with Precision in SAMSON
Exporting Ligand Trajectories for Reaction Coordinate Studies in SAMSON
Tired of Unsupported Molecular File Formats? Here’s How SAMSON Can Help
Visualizing the Spike Protein’s Dance: A Molecular Trajectory for Modelers
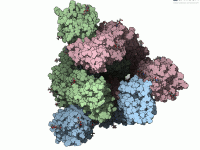
For molecular modelers working on viral proteins, computing conformational transitions between known structures is a frequent challenge. Whether designing potential inhibitors, studying viral entry, or simply trying to understand structural dynamics, one practical problem remains: how to interpolate between two…