Save Time and Generate Molecular Scripts with Natural Language in SAMSON
Making Sense of PMF with WHAM in SAMSON’s GROMACS Wizard
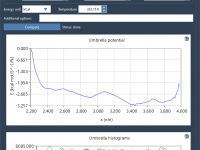
If you’ve ever run umbrella sampling simulations and found yourself stuck at the next step—extracting the Potential of Mean Force (PMF)—you’re not alone. PMF profiles help interpret free energy landscapes, crucial in understanding molecular interactions, binding affinities, and conformational changes.…