Author: OneAngstrom
Save Time and Improve Molecular Presentations with Visual Presets in SAMSON
Extending Molecular Modeling Tools for Your Research Needs
Filtering Molecular Models by Partial Charge Using NSL
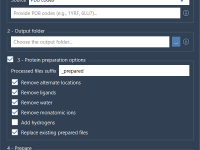
Cleaning 100 PDBs in One Go: Efficient Batch Protein Preparation with SAMSON
Quickly Find Conformations by Atom Count in SAMSON
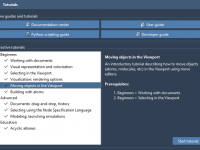
Struggling to Learn Molecular Modeling Tools? SAMSON’s Built-In Tutorials Make It Easier
How Symmetry Detection Can Minimize Simulation Time
Getting Started with Geometry Optimization in SAMSON Using the FIRE Minimizer

Preparing a molecular model for simulation often involves an essential but sometimes time-consuming step: geometry optimization. When atoms are not in energetically favorable positions, your simulation might diverge or waste computational resources on fixing geometry-related problems. Enter the FIRE Minimizer,…