Author: OneAngstrom
Struggling to Select Residues in 3D? Try This View Instead.
Avoid Simulation Errors: How to Properly Set Up Protein-Ligand Models in SAMSON
Before running advanced analyses such as ligand unbinding pathway searches or energy minimizations, it’s critical to properly prepare your molecular models. For many molecular modelers, skipping this step—or not doing it thoroughly—leads to frustrating errors during simulation, non-physical results, or…
Avoid Broken Pipelines: Choosing the Right Input for NVT Equilibration in GROMACS Wizard
Making Time Stand Still in Molecular Animations
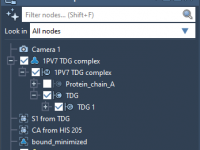
Tired of Losing Track of Your Molecules? Master the SAMSON Document View
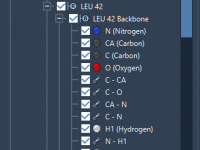
As molecular modeling projects grow in complexity, keeping everything organized—especially when dealing with multiple structures, simulations, and annotations—can become a real challenge. Mistakenly overwriting an important molecule, struggling to recall how a selection was made, or navigating through scattered files…
Add Subtle Motion to Molecular Visualizations with the Rock Animation in SAMSON
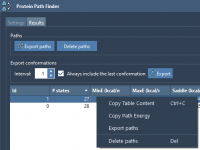
Practical Tips to Export Protein Conformational Pathways in SAMSON
A Better Way to Move Molecules: Using the Local Move Editor in SAMSON
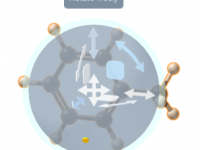
When modeling complex molecular systems, precise manipulation of structures is key. Whether you’re arranging ligands, adjusting dihedrals, or preparing a visual output, simple grab-and-drag operations often aren’t enough. Many modelers struggle with getting molecules to move exactly how they want—often…