Author: OneAngstrom
How to Align Specific Regions of Proteins in SAMSON
Avoid This Mistake When Defining the Simulation Box for COM Pulling
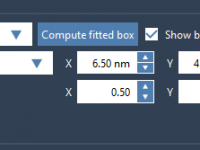
When setting up molecular dynamics (MD) simulations with pulling forces—especially center-of-mass (COM) pulling—one common but potentially problematic decision is defining the simulation box size. Selecting inappropriate box dimensions can interfere with the pulling process and compromise the reliability of results.…
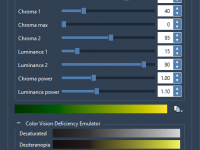
Choosing the Right Color Palette in Molecular Modeling
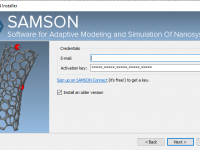
Need an Older Version of SAMSON? Here’s How to Install It Properly
Struggling with Molecular Data Export? Here’s What You Can Do in SAMSON
Need an Older Version of SAMSON? Here’s the Right Way to Do It
How to Select Molecules by Substructure without Losing Your Place
One of the recurring challenges for molecular modelers working with SMILES files is identifying and isolating molecules that contain specific substructures. Searching by substructure matters—a lot—when you’re building small molecule libraries, designing focused chemical series, or preparing datasets for further…
Handling Molecular Trajectories with the Play Path Animation in SAMSON
Working with conformational pathways and molecular dynamics trajectories often requires efficient ways to visualize movement between structures or along predefined paths. Molecular modelers frequently face this need, whether to analyze results, showcase simulations, or communicate structural changes over time. To…