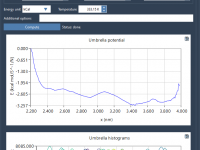
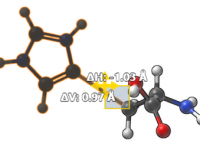
Mastering Molecular Visuals with SAMSON’s Color Schemes
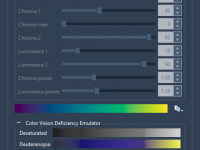
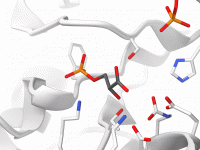
For molecular modelers and researchers, accurately visualizing molecular structures is not just essential; it can be transformative. The ability to spot patterns, recognize structural details, and communicate insights effectively often depends on how well molecular representations are highlighted. One of…