Author: OneAngstrom
Mastering the Pause Animation in Molecular Presentations
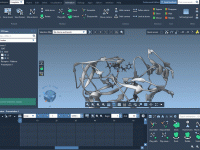
How to Build Lipid Layers Around Proteins with SAMSON’s Molecular Box Builder
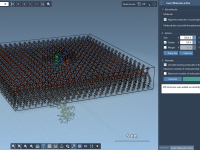
For molecular modelers involved in studying membrane proteins or simulating lipid-protein interactions, creating lipid bilayers around proteins can be a labor-intensive task. This process often involves multiple tools, some guesswork, and significant manual intervention. However, with SAMSON’s Molecular Box Builder,…
Streamline Molecule Design with Fragment Replacement
Master Molecular Modeling with Interactive Tutorials in SAMSON
Simplifying Molecular Animations: Undocking with SAMSON
Simplify Constrained Simulations with the Simulate Animation in SAMSON.
Molecular modeling often involves intricate simulations where systems must adhere to specific constraints or dynamically evolve based on several interacting factors. Conducting such simulations can sometimes feel daunting, especially when you need precise control over multiple elements. Enter the Simulate…
Streamlining Molecular Modeling with Property Model Attributes
For molecular modelers, managing the complexities of molecular structures and associated properties can be a daunting task. Documenting key attributes, improving visibility, and organizing data more effectively are all challenges that can significantly slow down workflows. Fortunately, property model attributes…