Author: OneAngstrom
Combining Monomer Sequences to Build Complex Polymers in SAMSON
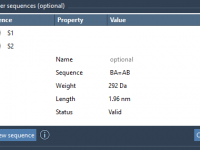
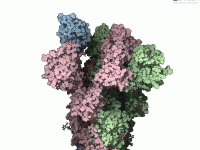
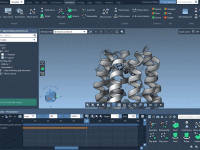
Designing complex polymers with repeating sequences can be challenging, especially when you need precise control over structure and connectivity. Whether you’re crafting custom scaffolds, developing synthetic chains, or simulating biopolymers, you often run into limitations with traditional modeling tools. This…