Author: OneAngstrom
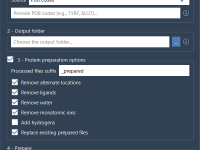
Cleaning Hundreds of Protein Structures with One Tool
Navigating Complex Molecular Documents in SAMSON: A Visual Guide to the Document View
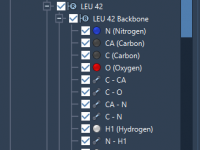
For molecular modelers dealing with multi-component systems, navigating and organizing complex structures can be time-consuming and error-prone. Whether you’re working with large protein complexes, nucleic acids, or synthetic compounds, keeping control over your molecular hierarchy is key to productivity —…
A Faster Way to Clean Up Molecular Visuals in SAMSON
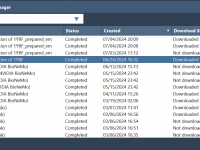
Can’t Keep Track of Your Cloud Simulations? Meet the SAMSON Job Manager
Make Atoms Vanish in Style: A Practical Guide to Concealment Animations in Molecular Scenes
Creating molecular presentations that feel clean and professional takes more than accurate models—it requires thoughtful visual storytelling. One common challenge for molecular modelers and educators alike is managing visual complexity in dense molecular scenes. Sometimes, key structures are hidden behind…
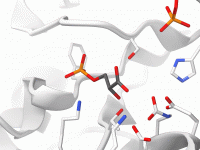
Improving Ligand Unbinding Pathways with the P-NEB Method in SAMSON
For molecular modelers working on protein-ligand interactions, refining transition pathways between binding and unbinding states is essential—but often challenging. Whether you’re studying conformational changes, binding affinity, or energy landscapes, having a more precise transition path between low-energy states directly improves…
Naming and Selecting Cameras in SAMSON’s Node Specification Language
Choosing the Right Color Palette in Molecular Visualizations: Why It Matters
Choosing the Right Visual Models in SAMSON: A Practical Guide for Molecular Modelers

As a molecular modeler, visualizing structures effectively is vital—not just for presentation purposes, but also for developing an intuitive understanding of complex nanosystems. Whether you’re analyzing protein folding, inspecting electrostatic fields, or preparing publication-quality visuals, selecting an appropriate visual model…