Author: OneAngstrom
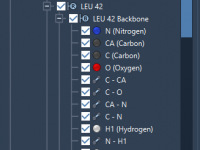
Simplifying Molecular Data Management with File Attributes
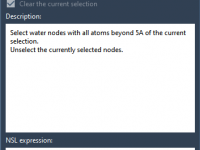
Deleting Crystal Waters Outside the Active Site for Accurate Simulations
Mastering Molecular Patterns with SAMSON’s Editors
Molecular modeling often involves repetitive tasks, such as creating patterns of atoms or molecules for various applications in nanotechnology, biomolecular modeling, or material science. Replicating molecular structures manually can be tedious, especially when dealing with hundreds of thousands of atoms.…
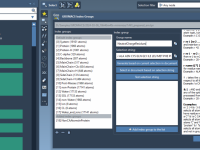
Simplify Molecular Modeling with Custom Index Groups in SAMSON
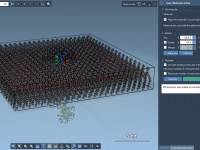
A Step-by-Step Guide to Building Lipid Layers Around Proteins
Essential Tips for Defining a Simulation Box in GROMACS Wizard
Mastering Zoom Camera Animations for Molecular Modeling
Simplifying Molecular Structure Optimization with Interactive Minimization in SAMSON
Molecular optimization is a critical step in computational chemistry, helping researchers refine structures for improved accuracy in simulations and analyses. However, achieving efficient and precise optimization can sometimes feel daunting. That’s where the interactive minimization capabilities of SAMSON can provide…