Author: OneAngstrom
Simplifying Molecular Structure Optimization with Interactive Minimization in SAMSON
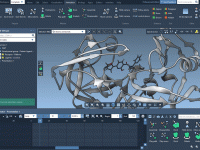
Molecular optimization is a critical step in computational chemistry, helping researchers refine structures for improved accuracy in simulations and analyses. However, achieving efficient and precise optimization can sometimes feel daunting. That’s where the interactive minimization capabilities of SAMSON can provide…
Preparing a Coarse-Grained System with GROMACS Wizard
Streamline Molecular Transparency with the Appear Animation in SAMSON
Molecular modeling often involves dynamic presentations to communicate key insights, an especially crucial task for researchers aiming to depict structural changes, interactions, or mechanistic pathways. One subtle but vital element in such visual storytelling is the progressive revelation of elements…
Mastering Python Package Management in SAMSON
Streamlining Substructure Searches with the SMILES Manager in SAMSON.
Streamlining Protein-Ligand Docking with the FITTED Suite
Protein-ligand docking is a crucial task in molecular modeling, aiding researchers in drug discovery and understanding molecular interactions. However, the process is often complex, involving intricate system preparations, docking parameter configurations, and optimization steps. If you’re looking for a way…