Author: OneAngstrom
Mastering Molecular Trajectories with the Play Path Animation in SAMSON
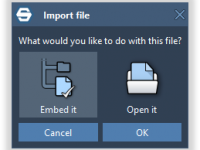
Seamlessly Embedding Files and Data for Molecular Design Projects
Simplify Molecular Trajectory Visualization with Play Path Animation
Understanding Visibility Control in Molecular Modeling
Accelerating Molecular Geometry Optimization with FIRE.
Simplify Carbon Nanotube Modeling Using the Nanotube Creator Tool

Modeling carbon nanotubes (CNTs) can be a complex and time-intensive task for molecular designers. Whether you’re designing nanodevices or studying the mechanical properties of CNTs, achieving precision and efficiency is paramount. Fortunately, SAMSON’s Nanotube Creator Extension is a powerful tool…
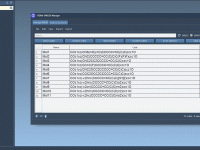
Effortlessly Generate 3D Structures from SMILES Strings with the SAMSON SMILES Manager
Understanding Residue Polarity in Molecular Modeling
Molecular modelers frequently need to work with residues that exhibit specific polarities for their research or simulations. Whether modeling protein-ligand interactions or analyzing molecular structures, knowing how to filter residues based on their polarity can significantly enhance productivity and precision.…