Author: OneAngstrom
Mastering Path Recording in Molecular Animations
For molecular modelers striving to capture complex atomic trajectories in their animations, SAMSON provides an intuitive and powerful feature: the Record path animation. This tool helps users record atoms’ movements during molecular presentations, effectively visualizing intricate interactions in a clear…
Demystifying Side Chain Attributes in Molecular Modeling
Efficiently Docking Ligand Libraries Using AutoDock Vina Extended
Bringing Molecular Visibility to Life with Flash Animation in SAMSON
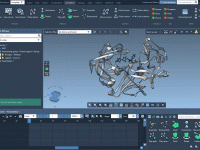
If you are a molecular modeler, you’ve likely encountered the need to highlight specific molecular nodes or sequences during your animation without overcomplicating the visualization. Whether for presentations, research insights, or interactive storytelling, an animation that controls when nodes appear…
Mastering Progressive Transparency with the Appear Animation in SAMSON
Tips for Filtering Structural Models Using SAMSON Attributes
When working on molecular modeling projects, quickly isolating and analyzing specific structural models can streamline workflows and enhance productivity. SAMSON, the integrative molecular design platform, makes this process intuitive through its robust set of structuralModel attributes. These attributes allow modelers…
Navigating Molecule Attributes in SAMSON’s Node Specification Language
Mastering Smooth Rotations with the Orbit Camera Tool in SAMSON
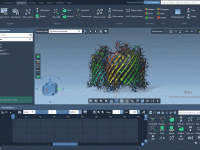
Molecular modeling often revolves around effectively presenting and analyzing molecular structures. One common challenge is creating smooth, dynamic camera rotations to highlight specific features, especially during collaborative presentations or while capturing animations for research documentation. SAMSON’s Orbit camera animation tool…