Effortlessly Managing Molecular Documents in SAMSON
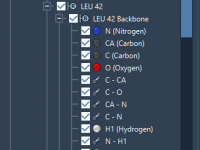
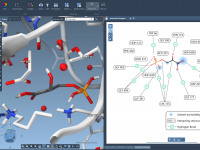
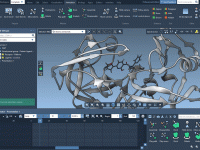
Molecular modelers often face the daunting task of organizing intricate structures, scripts, and datasets efficiently. Managing molecular documents can become increasingly complex when juggling multiple molecules or configurations. The good news? SAMSON, the integrative molecular design platform, offers a structured…