Author: OneAngstrom
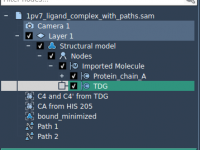
How to Select Chains by Atom, Residue, or Segment Counts in SAMSON
When exploring complex biomolecular systems, being able to efficiently select and manipulate specific chain structures is essential for insightful analysis. Whether you’re visualizing protein assemblies, preparing systems for simulation, or filtering out unwanted models, having control over chain-level attributes can…
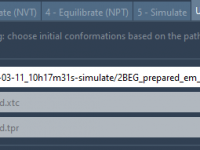
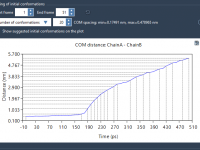
Selecting Initial Conformations for Umbrella Sampling Without the Headache
Choosing Effective Color Palettes for Molecular Visualizations
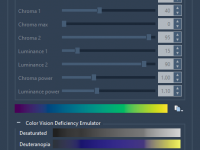
Molecular visualizations communicate complex structural information. But if you’ve ever found your protein coloring scheme hard to interpret—or worse, misleading—you’re not alone. Poor color choice can obscure relevant features, strain the eyes, or confuse colleagues with color vision deficiencies. Fortunately,…
Achieving Realistic Molecular Visuals with Material Presets in SAMSON
Need an Earlier Version of SAMSON? Here’s How to Get It
Working with DNA Origami in SAMSON: Simplifying Complex Nanostructures
Designing and simulating DNA origami structures has become an increasingly important part of molecular modeling and nanotechnology research. Whether you’re designing nanorobots, cages for drug delivery, or programmable matter, one common frustration is the challenge of managing and visualizing complex…
Finding the Right Look: Discrete Color Palettes in SAMSON
Visualizing Atomic Regions with Mathematical Expressions

When working with complex molecular systems, it’s often important to isolate specific atom groups for visualization or modification. Yet, selecting atoms manually can become tedious and error-prone, especially with large systems like crystals, biomolecules, or nanostructures. SAMSON’s Atoms Selector Extension…