Author: OneAngstrom
Progressive Disappearance in Molecular Models: A Clearer Way to Present Complex Structures
When Less is More: Concealing Atoms to Clarify Molecular Mechanisms
Avoiding Input Errors in GROMACS Equilibration: A Quick Guide to Selecting Structures
Making Fragments Fit: A Closer Look at Orienting Molecular Fragments in SAMSON
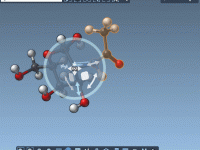
When building complex molecular models, one of the recurring challenges is aligning functional groups or fragments precisely and efficiently. Incorrect orientation can result in steric clashes, unrealistic geometries, or simply more work adjusting everything afterwards. If you’ve ever tried rotating…
Tweaking the Universal Force Field: Customizing Typization in SAMSON
Model Fade In, Fade Out: Animating Molecular Structures with Pulse
Effortlessly Navigate Molecular Design Changes with Undo and Redo
Quickly Find and Hide Labels in SAMSON Using NSL
Tired of Static Molecular Presentations? Here’s a Simple Way to Add Motion
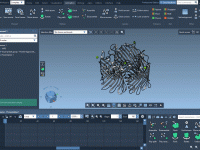
When trying to communicate molecular structures and processes effectively, nothing conveys spatial relationships and dynamics better than motion. Yet, many molecular modelers still present their work using static snapshots, missing the opportunity to make insights more intuitive and memorable. Fortunately,…