Tired of Static Molecular Presentations? Here’s a Simple Way to Add Motion
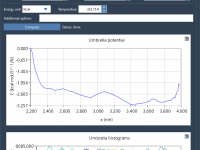
When trying to communicate molecular structures and processes effectively, nothing conveys spatial relationships and dynamics better than motion. Yet, many molecular modelers still present their work using static snapshots, missing the opportunity to make insights more intuitive and memorable. Fortunately,…