Author: OneAngstrom
Making Molecular Visualization Simpler: Understanding the ‘Hidden’ Attribute in SAMSON
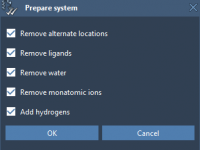
Streamlining Protein Preparation for Docking Simulations
Understanding Render Preset Attributes for Precise Molecular Visualization
For molecular modelers, a common challenge lies in the accurate and efficient visualization of molecular systems. Effective visualization often requires fine-tuning the rendering settings to highlight specific structures or properties. This is exactly where render preset attributes in SAMSON’s Node…
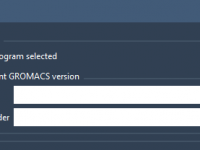
Customizing your GROMACS version in SAMSON for better reproducibility.
Understand and Use Conformation Attributes for Molecular Modeling
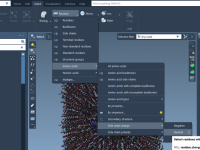
Mastering Visual Filtering with Visibility Flags in SAMSON
Mastering Background Transitions in SAMSON Presentations
Creating engaging and polished molecular presentations is essential for effectively communicating complex ideas. For those who rely on SAMSON’s molecular design platform, the Set Background animation provides a powerful way to elevate storytelling by seamlessly transitioning between backgrounds during a…