Understanding and Using Chain Attributes in Molecular Modeling
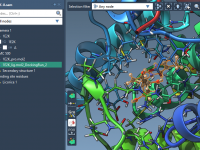
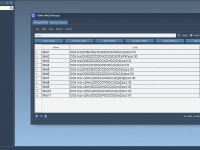
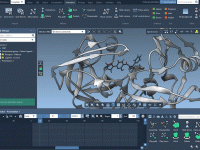
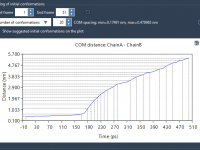
Molecular modeling often involves navigating complex datasets to precisely identify and manipulate molecular chains in your systems. When using SAMSON’s integrative molecular design platform, the proper understanding of “chain attributes”—an important feature of its Node Specification Language (NSL)—can simplify this…