Author: OneAngstrom
Building Lipid Layers Around Proteins for Molecular Modeling
Streamlining DNA Nanostructure Imports with Adenita.
For molecular modelers working with advanced DNA nanostructures, transferring designs between different tools can be a time-intensive task. Whether you’re leveraging designs from Cadnano or incorporating previously-created components, ensuring a smooth import can save hours of manual rework. This is…
Simplify Your Molecular Modeling Workflow with SAMSON Extensions.
Simplify Protein Conformational Transitions with ARAP Interpolation
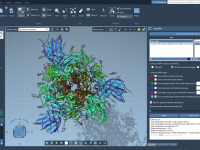
Molecular modelers often face significant challenges when analyzing protein conformational transitions. Whether you’re preparing reaction coordinates for free energy simulations or studying functional dynamics, generating smooth and realistic transition paths between protein structures can be daunting. The As-Rigid-As-Possible (ARAP) Interpolator…
Crafting a Strong Public Profile on SAMSON Connect
Choose the Perfect Discrete Color Palette for Molecular Modeling in SAMSON.
Creating compelling molecular visualizations is often a challenge for molecular modelers. Representing complex structures and interactions in an intuitive and visually appealing way can enhance communication, understanding, and even the modeling process itself. One critical component is choosing the right…
Accelerate Biomolecular Discoveries with AlphaFold-2 in SAMSON
For molecular modelers seeking to unravel the complexities of biomolecular structures, accurate prediction tools are an integral part of the workflow. Tedious manual modeling processes and limited access to advanced computational resources often create bottlenecks. SAMSON offers a powerful solution…