Author: OneAngstrom
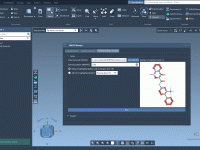
Swapping Atoms, Saving Time: Quick Analogue Generation in SAMSON
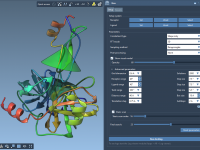
FASTA In, Structure Out: Predicting Proteins with AlphaFold-2 in SAMSON
Predicting protein structures used to be a long, resource-heavy process, often requiring local installations or complex data preparation. Today, especially with the introduction of AlphaFold-2, molecular modelers have access to powerful prediction engines remotely. However, integrating these capabilities into real…
Reducing False Positives in Protein Docking with Smart Search Domain Setup
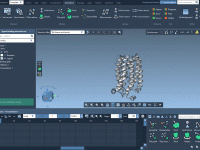
Making Molecular Trajectories Understandable with the Play Path Animation
Finding the Right Groups in SAMSON Using Node Group Attributes
Struggling with molecular file formats? Here’s how SAMSON handles it.
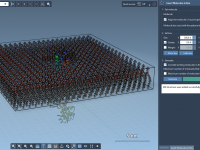
Placing Lipids Around Membrane Proteins Without Tears or Scripts
Want to Track How a Ligand Unbinds? Try This Simple Visualization
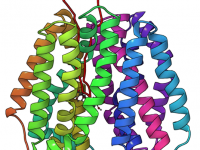
One of the recurring challenges in molecular modeling is making sense of complex atomic trajectories. Whether studying how a ligand unbinds from a protein or following large-scale conformational changes, analyzing raw coordinates can quickly become overwhelming. Fortunately, there’s a straightforward…