Author: OneAngstrom
A Simple Way to Build Carbon Nanotube Models in SAMSON
Building Custom Crystal Structures: A Step-by-Step Guide for Molecular Modelers

Creating custom crystal structures is a valuable skill for anyone working in materials science, solid-state chemistry, or molecular modeling. Whether you’re studying lattice strain, dopant behavior, or crystalline defects, simulating accurate, made-to-measure crystal models is essential. Fortunately, SAMSON’s Crystal Creator…
Why Having an Undo Button Matters More Than You Think in Molecular Modeling
Make Molecular Interactions Move: A Simple Way to Present Docking with Motion
Making Molecular Models More Expressive with the Rock Animation
Finding the Right Bonds: A Quick Guide to Filtering by Type in SAMSON’s NSL
What to do when your molecular file doesn’t load in SAMSON
A smoother way to navigate molecular paths
How to Create Custom Index Groups in GROMACS Without Writing NDX Files by Hand
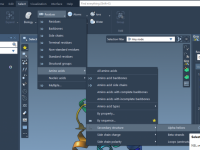
When running GROMACS simulations, defining custom selection groups—also known as index groups—can be essential. These groups allow for targeted analysis, pulling configurations, and interaction computations. However, editing GROMACS .ndx files manually is tedious and error-prone, especially when dealing with large…