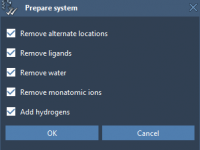
How to Quickly Find and Adjust Render Presets in SAMSON Models
Molecular modelers often spend a significant amount of time tweaking visual parameters to highlight structural features. Whether it’s for presentations, publications, or just better visual inspection, being able to identify and manipulate specific render presets becomes essential. In SAMSON, render…