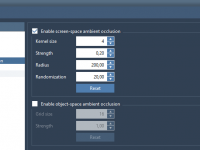
Filtering Residues by Charge in SAMSON: A Quick Guide for Molecular Modelers
When dealing with biomolecular systems, especially proteins, understanding the charge distribution of residues plays a critical role in many modeling tasks. Whether you’re preparing input for electrostatics calculations, identifying potential binding sites, or simply studying protein-ligand interactions, selecting residues based…
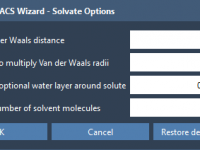
Avoiding Solvent Clashes in Coarse-Grained Simulations with GROMACS Wizard
Making Molecular Components Appear on Cue in Your Animations
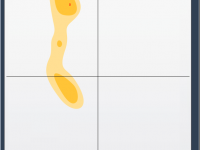
Clicking Your Way Through Proteins: Inspecting Residues with the Interactive Ramachandran Plot
Are You Modeling the Right Bond Type?
Making Your Molecules Stand Out: A Guided Tour of SAMSON’s Rendering Preferences
Replaying Molecular Trajectories in Reverse: A Timesaving Trick in SAMSON
Freeing Bound Molecules: Create Undocking Animations in SAMSON
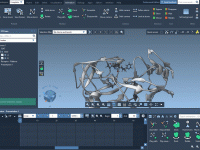
When modeling molecular interactions, demonstrating a ligand binding to a receptor is essential. But what about the reverse process? Undocking animations are just as important, especially when preparing presentations, educational content, or investigating energy release during the unbinding stage. In…
Never Guess Again: Measuring Distances Between Atoms in SAMSON
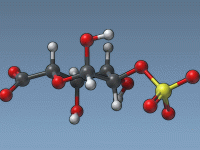
Whether you’re analyzing protein-ligand interactions or checking geometry optimization results, measuring precise atomic distances is a routine—but vital—task in molecular modeling. Yet, for many users, keeping track of those measurements or quickly accessing the most relevant ones can be frustrating…