Want Certain Molecules to Briefly Appear? Try the Flash Animation in SAMSON
Finding the Right Tools for Molecular Modeling in the SAMSON Marketplace
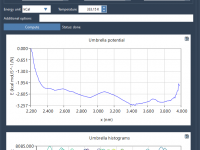
Conformations vs. Paths: A Time-Saving Tip When Using P-NEB
Before You Plan Protein Transitions, Minimize First — Here’s How
Constrained Simulations in SAMSON: Ensuring Realistic Molecular Motions
Building a Lipid Layer Around a Protein Without Manual Guesswork
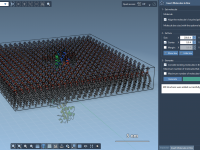
Preparing membrane-protein systems for simulation is a task that molecular modelers regularly face, especially when modeling transmembrane proteins. However, manually inserting lipids and ensuring proper alignment around a protein can be time-consuming and error-prone. If you’ve ever tried to construct…