Freezing Atoms in Time: A Simple Technique for Clearer Molecular Animations
A simple way to move your molecular view sideways
Making Molecular Models Appear and Vanish: A Simple Trick with Flash Animation in SAMSON
Installing Python Packages in SAMSON—What Molecular Modelers Need to Know
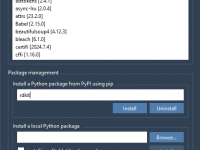
When using SAMSON for molecular modeling, integrating powerful Python packages can significantly improve your workflow, especially if you’re automating simulations, analyzing data, or integrating machine learning into your research. However, choosing the right environment and knowing how to manage Python…