How to Quickly Filter Cameras in Large Molecular Projects
Avoid Topology Errors in MARTINI Coarse-Grained Models of Protein Replicas
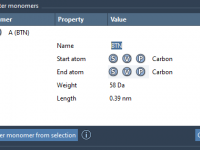
When setting up coarse-grained (CG) molecular dynamics simulations using the MARTINI force field, one recurring roadblock for molecular modelers is handling multiple replicas of the same protein. Whether you’re modeling crowded cellular environments or testing multimeric assemblies, creating multiple instances…