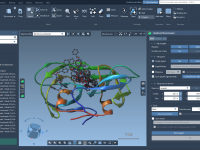
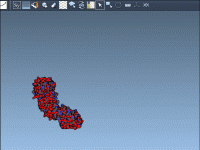
Better Molecular Presentations: Using Background Transitions in SAMSON
Molecular modelers often need to communicate complex information through presentations. Whether you’re presenting at a conference, teaching a class, or preparing materials for collaborators, clarity and visual impact matter. But switching between different molecular scenes can be visually jarring. A…