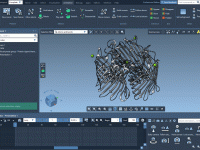
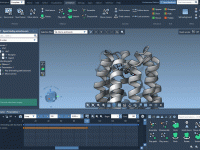
Never Lose Sight of Key Molecules During Simulations
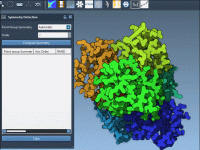
Choosing the Right Symmetry Group in Large Molecular Assemblies
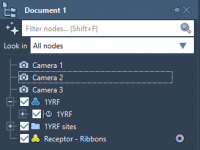
Switching Views Instantly: Using Multiple Cameras in SAMSON
Create Slide-Like Molecular Presentations in SAMSON with the Stop Animation
Tired of Managing Molecular Simulations Locally? Try This Alternative
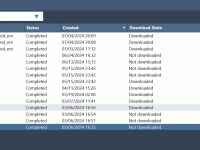
Running complex molecular modeling jobs—such as protein structure prediction or molecular dynamics simulations—often requires substantial computing power, software installations, and careful monitoring of progress. For researchers dealing with local system limits, slow workflows, or collaboration headaches, SAMSON’s cloud-based Job Manager…
Why Did My Labels Disappear? Understanding Label Visibility in SAMSON
Indexing Beyond the Defaults: Smarter GROMACS Simulations with Custom Groups
From Repetition to Pattern: Designing Monomer Sequences with Polymer Builder
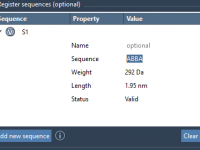
Building custom polymers can quickly become complex—especially when you’re not just connecting one identical monomer after another, but working with patterned sequences. Many computational chemists and molecular designers struggle with modeling repeat units that reflect experimental conditions or functional motifs.…
Filtering Molecular Models by Partial Charge in SAMSON
When working with complex molecular systems, identifying specific structural groups based on electrostatic properties can be essential. Whether you’re screening ligands, analyzing biomolecular systems, or designing new compounds, partial charges help determine how molecules interact. Yet, combing through thousands of…