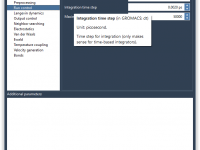
Running GROMACS Simulations in the Cloud Without Installing GROMACS
Quickly Animate Atomic Motion Using Keyframes in SAMSON
Creating effective molecular animations can be time-consuming, especially when you’re trying to convey subtle structural changes or transformations in a clear and controlled manner. Whether you’re preparing a scientific presentation or building an educational visualization, you’ve probably found yourself wrestling…
Speed Up Your Molecular Visualizations with Custom Visual Presets
Creating clear and expressive molecular visualizations can be a time-consuming task. Whether you’re working on a presentation, preparing figures for publications, or simply exploring a structure, it often involves repetitive steps: selecting atoms, applying representations, choosing color schemes, adjusting views,…