From Atoms to Lattices: Easily Create Custom Crystals in SAMSON
Minimizing Only Part of a Molecule in SAMSON Without Affecting the Rest
One common challenge in molecular modeling is selectively optimizing only certain parts of a molecular structure while leaving the rest untouched. This becomes particularly relevant when working on large biomolecules, designing ligands, or when assessing localized conformational changes. Minimizing an…
Quickly Setting Up Protein Docking in SAMSON with Hex
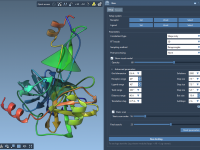
Manually configuring a protein docking setup can be tedious and error-prone, especially when dealing with complex biomolecular structures. One common difficulty for molecular modelers is quickly and accurately preparing a system for docking simulations while minimizing preprocessing time. Fortunately, SAMSON…
Tired of Writing Custom Parsers? How Importers Can Save You Hours
Quickly Find Molecular Subsets in SAMSON with Folder Attribute Filters
From Chaos to Structure: Easily Animate Molecular Assembly
Animating Molecular Pulses: A Simple Way to Clarify Transient Structures
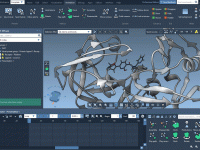
In molecular modeling, one persistent challenge is effectively conveying how structures transiently appear and disappear during processes like docking, folding, or dynamic transformations. Researchers often want to emphasize certain regions of a molecular system momentarily — perhaps to draw attention…