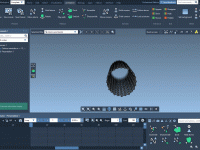
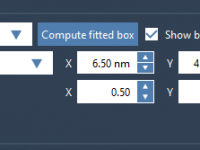
Planning Your Simulation Space: How to Define the Periodic Box for COM Pulling in GROMACS Wizard
Getting Depth Right: An Easy Rendering Trick for Molecular Models
Quickly Find and Filter Your Cameras in SAMSON with NSL
Choosing Better Discrete Color Palettes for Molecular Visualizations in SAMSON
Camera Movements Made Simple: Orbit, Zoom, Dolly and More in SAMSON
Showing Only What Matters in Molecular Presentations
A Simple Way to Show Molecular Assembly in Motion
Making Vertical Camera Movements Easier in Molecular Animations
Easily Find Selected Folders in Your Molecular Simulations
When conducting molecular simulations or organizing structural data, it’s common to work with complex models that contain numerous folders and nodes. A recurring challenge for molecular modelers is identifying which elements in a hierarchical structure are selected—especially after interacting with…