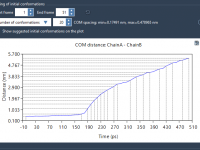
Running Molecular Simulations from Multiple Conformations without Repeating Yourself
Drawing Nanotubes with Your Mouse: A Faster Path to Carbon Nanotube Models

Creating molecular models can be a meticulous process—even more so when working with nanostructures like carbon nanotubes (CNTs). Whether you’re designing nanoscale sensors, exploring molecular transport systems, or preparing systems for simulation, setting up your model quickly and accurately matters.…
Making Molecules Move: A Closer Look at ‘Move atoms’ in SAMSON
Quickly Select Specific Bonds in SAMSON Using NSL
When working on complex molecular systems, selectively targeting and analyzing specific kinds of bonds—by type, order, or length—can significantly speed up your workflow. Whether you’re reviewing molecular geometries, preparing simulations, or visualizing particular features, manually clicking through bonds or writing…