When Steepest Descent Isn’t Enough: A Simple Way to Speed Up Geometry Optimization with FIRE
Choosing Discrete Color Palettes That Make Molecular Models Clearer
How to Quickly Select and Filter Cameras in SAMSON Using NSL
Working with multiple cameras inside a complex molecular system can feel overwhelming. In integrative modeling platforms like SAMSON, cameras help visualize the system from different perspectives—especially useful during animation, scene capturing, or scientific reporting. However, selecting and managing these cameras…
Saving Time with Custom Index Groups in GROMACS Wizard
Bringing Molecules Into Focus: Customizing Rendering Preferences in SAMSON
Quickly Managing Label Visibility in Molecular Models
Making NVT Equilibration Easy: Automatically Load Inputs in SAMSON’s GROMACS Wizard
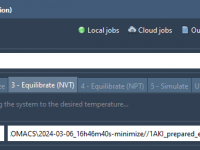
One of the recurring pain points for computational molecular scientists is managing the multiple input and output files between different simulation steps. This becomes especially cumbersome when performing sequential operations like energy minimization followed by NVT and NPT equilibrations, where…