An Easier Way to Build Multi-Walled Carbon Nanotubes for Simulations

Creating realistic models of complex nanostructures, such as multi-walled carbon nanotubes (MWCNTs), can be a time-consuming challenge for molecular modelers. Whether you’re exploring molecular transport, testing drug delivery systems, or simulating electronic properties, manually building these nanotubes atom-by-atom is impractical.…
Which Symmetry Group Should You Choose in Large Protein Assemblies?
Quickly Show or Hide Molecular Presentations with NSL Filters
Working on complex biomolecular systems often requires navigating through dozens — sometimes hundreds — of graphical elements: molecules, representations, overlays, labels, and more. When viewing becomes cluttered, productivity and clarity drop. That’s why visibility control is essential in molecular modeling…
Pausing Molecular Storytelling with Precision: Using the Stop Animation in SAMSON
When presenting complex molecular mechanisms, clarity and pacing matter. Introducing a new structure, showcasing an interaction, or highlighting a conformational change requires timing that matches your narrative—especially when explaining concepts to students, collaborators, or during scientific talks. That’s where the…
Need to Move Your Molecular View Vertically? Try the Pedestal Camera Effect
Fine-tuning Molecules in SAMSON: How to Quickly Edit and Customize Analogs
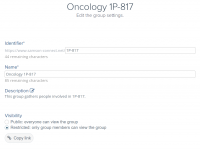
Quickly Select and Control Chain Visibility in Molecular Models
When working with complex biomolecular systems, it’s common to navigate and manipulate multiple chains alongside other structural groups. Whether you’re preparing publication-quality visuals, analyzing specific interactions, or just trying to declutter your scene, being able to selectively show or hide…