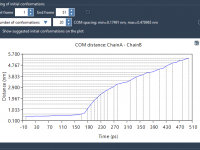
Rotate and Align Molecules Precisely in SAMSON Using Move Editors
Quickly Select Molecular Paths with More Than 100 Atoms in SAMSON
Avoiding Solvent Clashes in Coarse-Grained Systems: A Common GROMACS Pitfall
Finding Aromatic Atoms Quickly in Molecular Models with NSL
How to filter molecular structures by atom count in SAMSON
A Simple Trick to Showcase Molecular Motion with the Rock Animation
For molecular modelers, visually communicating dynamic behavior is essential—whether it’s for presentations, publications, or team discussions. Yet, it’s often difficult to convey subtle molecular motions effectively and understandably. One recurring challenge? Showing how groups of particles move together in a…
Avoid Rework: How to Auto-Fill Input Files for NVT Equilibration in SAMSON
Using NSL to Control Note Visibility in SAMSON
Creating Smooth Protein Transition Paths with ARAP Interpolation
When modeling molecular processes, capturing transitions between different conformations of a protein is often essential. Whether you’re exploring activation pathways, refining docking poses, or setting up umbrella sampling simulations, intermediate structures along these conformational paths are a valuable resource. But…