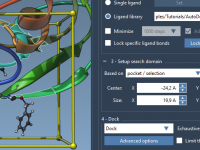
Quickly Separate Molecular Components for Clear Presentations in SAMSON
Moving the Camera, Not the Molecule: A Simple Way to Enhance Molecular Presentations

When preparing molecular animations for presentations or scientific communication, molecular modelers often face a recurring challenge: how to present the structure dynamically without altering its real position? Sometimes, rotating or moving the molecule can be misleading or distracting—especially when showing…
From SMILES to 3D Structures in Seconds: A Practical Guide
Matching Chains: How to Align Protein Sequences by Chain for Better Comparative Modeling
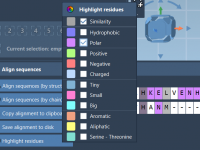
Accurately comparing proteins is a cornerstone of molecular modeling, whether you’re examining point mutations, comparing homologs across species, or preparing a structure for homology modeling. But when working with multi-chain proteins or oligomeric complexes, sequence alignment at the chain level…
Adding and Removing SAMSON Extensions: What Molecular Modelers Need to Know
Why You Don’t Have to Code Everything From Scratch in Molecular Modeling
One of the recurring challenges in molecular modeling is that integrating external tools and scripts often disrupts your workflow. Whether you rely on docking programs, simulation pipelines, or visualization utilities, switching between environments—coupled with writing tedious wrapper code—can slow down…