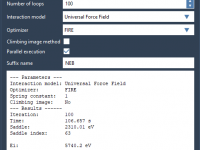
Enhancing Molecular Pathways with the P-NEB Method
Streamlining Molecular Animations with Visibility and Selection Tools
Animating molecular structures can be an essential part of molecular modeling workflows, whether you’re visualizing dynamics or generating intuitive presentations. Yet, managing the visibility and selection statuses of animation nodes can sometimes feel cumbersome. Fortunately, SAMSON’s Animation attribute space (short…
Simplify Molecular Dynamics Setup with the GROMACS Wizard in SAMSON
Molecular modelers often face the daunting challenge of setting up simulations efficiently. Preparing a system, ensuring compatibility with tools like GROMACS, and managing installations can be time-consuming and error-prone. If you’ve ever wished for a streamlined, user-friendly approach, the GROMACS…
Mastering Conformation Attributes in SAMSON for Molecular Design
For molecular modelers, navigating and managing molecular conformations effectively can often be a significant challenge. Whether you’re dealing with detailed structural analysis or large-scale molecular conformations, organizing and selecting relevant features is a frequent need. SAMSON provides a powerful solution…