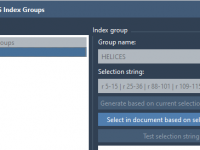
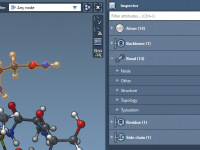
Save Time in GROMACS Setup by Creating Custom Atom Groups Visually
Quickly Find the Lights You’re Looking For: Making Sense of Visibility Flags
Quickly Filter and Manage Custom Visual Styles in SAMSON Using NSL
Making Specific Molecules Visible in Animations: A Quick Guide
Why the Sampling Box Matters When Exploring Ligand Unbinding Paths
Realigning Just the Part That Matters: Region-Specific Protein Alignment in SAMSON
Organize Your Molecular Data with the Document View in SAMSON
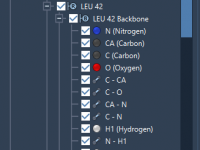
Keeping track of complex molecular models can be a challenge, especially when working with large biomolecular systems or managing multiple simulation setups. Whether you’re designing a drug candidate, investigating protein-ligand interactions, or comparing molecular structures, having a clear overview and…
Stop Guessing Atom Attributes: A Closer Look at the Inspector in SAMSON
Cutting Crystals with Miller Indices in SAMSON: A Quick Guide
For molecular modelers working with solid-state materials, understanding and manipulating crystal structures is essential. But cutting crystals along specific directions—especially when dealing with Miller indices—can be confusing or tedious in many software environments. If you’re using SAMSON, the integrative molecular…