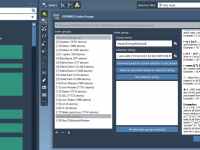
Cutting Crystals with Miller Indices in SAMSON: A Quick Guide
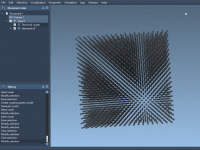
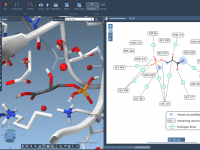
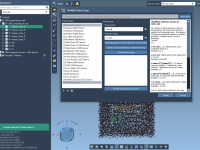
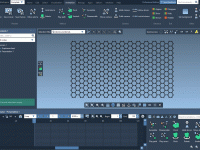
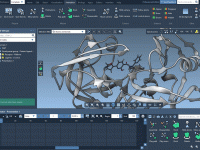
For molecular modelers working with solid-state materials, understanding and manipulating crystal structures is essential. But cutting crystals along specific directions—especially when dealing with Miller indices—can be confusing or tedious in many software environments. If you’re using SAMSON, the integrative molecular…